template class mrpt::graphslam::deciders::CLoopCloserERD¶
Edge Registration Decider scheme specialized in Loop Closing.
Current decider is implemented based on the following papers:
[1] Consistent Observation Grouping for Generating Metric-Topological Maps that Improves Robot Localization - J. Blanco, J. Gonzalez, J. Antonio Fernandez Madrigal, 2006
[2] Recognizing places using spectrally clustered local matches - E. Olson, 2009
Map type: 2D
MRPT rawlog format: #1, #2
Graph Type: CPosePDFGaussianInf
Observations: CObservation2DRangeScan
Edge Registration Strategy: ICP Scan-matching between nearby nodes
Loop Registration Strategy: Pair-wise Consistency of ICP Edges
The Loop closure registration strategy is described below:
We split the graph under-construction into groups of nodes. The groups are formatted based on the observations gathered in each node position. The actual split between the groups is decided by the minimum normalized Cut (minNcut) as described in [1].
Having assembled the groups of nodes, we find the groups that might contain loop closure edges (these groups contain successive nodes with large difference in their IDs). These groups are then split into two subgroups A, B, with the former containing the lower NodeIDs and the latter the higher. To minimize computational cost as well as the possibility of wrong loop closure registration, we search for loop closures only between the last nodes of group B and the first nodes of group A. Based on [2] the potential loop closure edges are not evaluated individually but rather in sets. Refer to [2] for insight on the algorithm and to evaluatePartitionsForLC method for the actual implementation. See below for images of potential Loop closure edges.
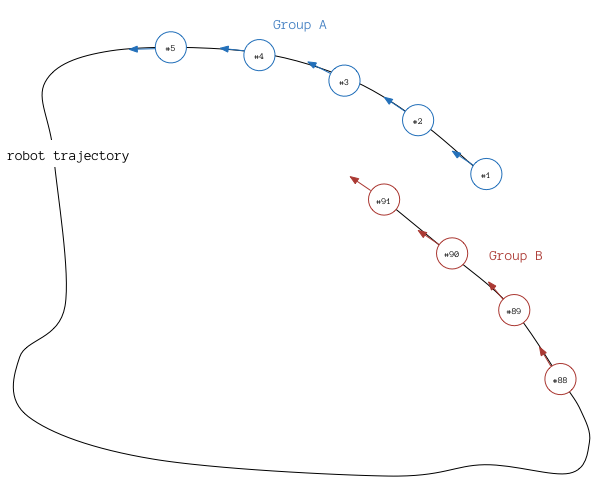
Loop closing schematic. blue nodes belong to group A and have lower NodeIDs, while red nodes belong to group B and have higher NodeIDs. Nodes of both groups A and B belong to the same partition based on their corresponding 2DRangeScan observations.
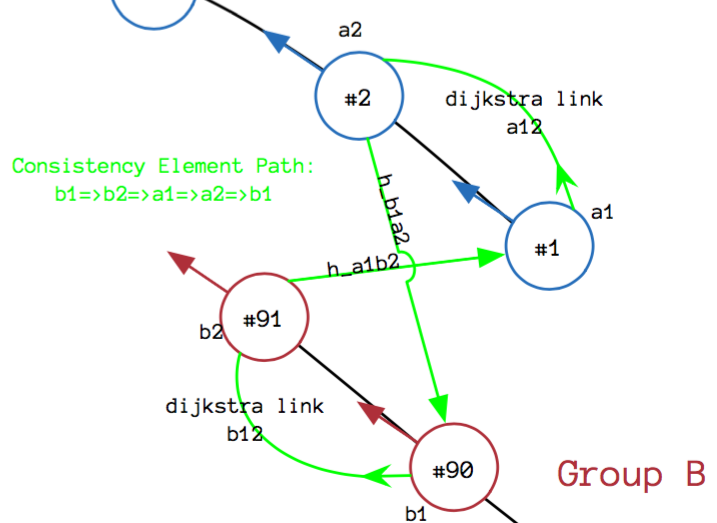
Rigid body transformation of hypotheses and two corresponding dijkstra links
Olson uses the following formula for evaluating the pairwise consistency between two hypotheses i,j:
\(A_{i,j} = e^{T \Sigma_T^{-1} T^T}\)
Where:
T is the rigid body transformation using the two hypotheses and the two Dijkstra Links connecting the nodes that form the hypotheses
\(\Sigma_T\) is the covariance matrix of the aforementioned rigid body transformation
However this formula is inconsistent with the rest of the paper explanations and mathematical formulas :
The author states that:
"This quantity is proportional to the probability density that the rigid-body transformation is the identity matrix (i.e., T = [0 0 0])"
This is inconsistent with the given formula. Suppose that a wrong loop closure is included in the \(A_{i,j}\), therefore the pairwise-consistency element should have a low value. For this to hold true the exponent of the consistency element shoud be small and neglecting the covariance matrix of rigid-body transformation (e.g. unit covariance matrix), \(T T^T\) should be small. When a wrong loop closure is evaluated the aforementioned quantity increases since the hypotheses do not form a correct loop. Therefore the worse the rigid-body transformation the higher the exponent term, therefore the higher the consistency element
Author uses the information matrix \(\Sigma_T^{-1}\) in the exponential. However in the optimal case (high certainty of two correct loop closure hypotheses) information matrix and rigid body transformation vector T have opposite effects in the exponent term:
\(\text{Correct loop closure} \Rightarrow T \rightarrow [0, 0, 0] \Rightarrow \text{exponent} \downarrow\)
\(\text{Correct loop closure} \Rightarrow \text{diagonal\_terms}(\Sigma_T^{-1}) \uparrow \Rightarrow \text{exponent} \uparrow\)
Based on the previous comments the following formula is used in the decider:
\(A_{i,j} = e^{-T \Sigma_T T^T}\)
class_verbosity
Section : EdgeRegistrationDeciderParameters
Default value : 1 (mrpt::system::LVL_INFO)
Required : FALSE
use_scan_matching
Section : EdgeRegistrationDeciderParameters
Default value : TRUE
Required : FALSE
Description :Indicates whether the decider uses scan matching between the current and previous laser scans to correct the robot trajectory.
visualize_laser_scans
Section : VisualizationParameters
Default value : 10
Required : FALSE
LC_min_nodeid_diff
Section : GeneralConfiguration
Default value : 30
Required : FALSE
Description : Minimum NodeID difference for an edge to be considered a loop closure.
LC_min_remote_nodes
Section : EdgeRegistrationDeciderParameters
Default value : 3
Required : FALSE
Description : Number of remote nodes that must exist in order for the Loop Closure procedure to commence
LC_eigenvalues_ratio_thresh
Section : EdgeRegistrationDeciderParameters
Default value : 2
Required : FALSE
Description : Minimum ratio of the two dominant eigenvalues for a loop closing hypotheses set to be considered valid
LC_check_curr_partition_only
Section : EdgeRegistrationDeciderParameters
Default value : TRUE
Required : FALSE
Description : Boolean flag indicating whether to check for loop closures only in the current node’s partition
visualize_map_partitions
Section : VisualizationParameters
Default value : TRUE
Required : FALSE
Class contains an instance of the mrpt::slam::CIncrementalMapPartitioner class and it parses the configuration parameters of the latter from the “EdgeRegistrationDeciderParameters” section. Refer to mrpt::slam::CIncrementalMapPartitioner documentation for its list of configuration parameters
See also:
mrpt::slam::CIncrementalMapPartitioner
#include <mrpt/graphslam/ERD/CLoopCloserERD.h> template <class GRAPH_T = typename mrpt::graphs::CNetworkOfPoses2DInf> class CLoopCloserERD: public mrpt::graphslam::deciders:: CRangeScanEdgeRegistrationDecider< GRAPH_T > { public: // typedefs typedef CRangeScanEdgeRegistrationDecider<GRAPH_T> parent_t; typedef typename GRAPH_T::constraint_t constraint_t; typedef typename GRAPH_T::constraint_t::type_value pose_t; typedef typename GRAPH_T::global_pose_t global_pose_t; typedef CLoopCloserERD<GRAPH_T> decider_t; typedef typename parent_t::range_ops_t range_ops_t; typedef typename parent_t::nodes_to_scans2D_t nodes_to_scans2D_t; typedef std::vector<std::vector<uint32_t>> partitions_t; typedef typename GRAPH_T::edges_map_t::const_iterator edges_citerator; typedef typename GRAPH_T::edges_map_t::iterator edges_iterator; typedef typename mrpt::graphs::detail::THypothesis<GRAPH_T> hypot_t; typedef std::vector<hypot_t> hypots_t; typedef std::vector<hypot_t*> hypotsp_t; typedef std::map<std::pair<hypot_t*, hypot_t*>, double> hypotsp_to_consist_t; typedef mrpt::graphslam::TUncertaintyPath<GRAPH_T> path_t; typedef std::vector<path_t> paths_t; typedef mrpt::graphslam::detail::TNodeProps<GRAPH_T> node_props_t; // structs struct TGenerateHypotsPoolAdParams; struct TGetICPEdgeAdParams; struct TLaserParams; struct TLoopClosureParams; // methods bool updateState( mrpt::obs::CActionCollection::Ptr action, mrpt::obs::CSensoryFrame::Ptr observations, mrpt::obs::CObservation::Ptr observation ); void setWindowManagerPtr(mrpt::graphslam::CWindowManager* win_manager); void notifyOfWindowEvents(const std::map<std::string, bool>& events_occurred); void getEdgesStats(std::map<std::string, int>* edge_types_to_num) const; void initializeVisuals(); void updateVisuals(); void loadParams(const std::string& source_fname); void printParams() const; void getDescriptiveReport(std::string* report_str) const; void getCurrentPartitions(partitions_t& partitions_out) const; const partitions_t& getCurrentPartitions() const; size_t getDijkstraExecutionThresh() const; void setDijkstraExecutionThresh(size_t new_thresh); void generateHypotsPool( const std::vector<uint32_t>& groupA, const std::vector<uint32_t>& groupB, hypotsp_t* generated_hypots, const TGenerateHypotsPoolAdParams* ad_params = nullptr ); void generatePWConsistenciesMatrix( const std::vector<uint32_t>& groupA, const std::vector<uint32_t>& groupB, const hypotsp_t& hypots_pool, mrpt::math::CMatrixDouble* consist_matrix, const paths_t* groupA_opt_paths = nullptr, const paths_t* groupB_opt_paths = nullptr ); void evalPWConsistenciesMatrix(const mrpt::math::CMatrixDouble& consist_matrix, const hypotsp_t& hypots_pool, hypotsp_t* valid_hypots); };
Typedefs¶
typedef CRangeScanEdgeRegistrationDecider<GRAPH_T> parent_t
Edge Registration Decider.
typedef CLoopCloserERD<GRAPH_T> decider_t
self type
Methods¶
size_t getDijkstraExecutionThresh() const
Return the minimum number of nodes that should exist in the graph prior to running Dijkstra.
void generateHypotsPool( const std::vector<uint32_t>& groupA, const std::vector<uint32_t>& groupB, hypotsp_t* generated_hypots, const TGenerateHypotsPoolAdParams* ad_params = nullptr )
Generate the hypothesis pool for all the inter-group constraints between two groups of nodes.
Parameters:
groupA |
First group to be tested |
groupB |
Second group to be tested |
generated_hypots |
Pool of generated hypothesis. Hypotheses are generated in the heap, so the caller is responsible of afterwards calling delete. |
void generatePWConsistenciesMatrix( const std::vector<uint32_t>& groupA, const std::vector<uint32_t>& groupB, const hypotsp_t& hypots_pool, mrpt::math::CMatrixDouble* consist_matrix, const paths_t* groupA_opt_paths = nullptr, const paths_t* groupB_opt_paths = nullptr )
Compute the pair-wise consistencies Matrix.
Parameters:
groupA |
First group to be used |
groupB |
Second group to be used |
hypots_pool |
Pool of hypothesis that has been generated between the two groups [out] consist_matrix Pointer to Pair-wise consistencies matrix that is to be filled |
groupA_opt_paths |
Pointer to vector of optimal paths that can be used instead of making queries to the m_node_optimal_paths class vector. See corresponding argument in generatePWConsistencyElement method |
groupB_opt_paths |
See also:
generatePWConsistencyElement
void evalPWConsistenciesMatrix( const mrpt::math::CMatrixDouble& consist_matrix, const hypotsp_t& hypots_pool, hypotsp_t* valid_hypots )
Evalute the consistencies matrix, fill the valid hypotheses.
Call to this method should be made right after generating the consistencies matrix using the generatePWConsistenciesMatrix method
See also: