Detailed Description
Functions | |
| double | mrpt::math::normalPDF (double x, double mu, double std) |
| Evaluates the univariate normal (Gaussian) distribution at a given point "x". More... | |
| template<class VECTORLIKE1 , class VECTORLIKE2 , class MATRIXLIKE > | |
| MATRIXLIKE::Scalar | mrpt::math::normalPDFInf (const VECTORLIKE1 &x, const VECTORLIKE2 &mu, const MATRIXLIKE &cov_inv, const bool scaled_pdf=false) |
| Evaluates the multivariate normal (Gaussian) distribution at a given point "x". More... | |
| template<class VECTORLIKE1 , class VECTORLIKE2 , class MATRIXLIKE > | |
| MATRIXLIKE::Scalar | mrpt::math::normalPDF (const VECTORLIKE1 &x, const VECTORLIKE2 &mu, const MATRIXLIKE &cov, const bool scaled_pdf=false) |
| Evaluates the multivariate normal (Gaussian) distribution at a given point "x". More... | |
| template<typename VECTORLIKE , typename MATRIXLIKE > | |
| MATRIXLIKE::Scalar | mrpt::math::normalPDF (const VECTORLIKE &d, const MATRIXLIKE &cov) |
| Evaluates the multivariate normal (Gaussian) distribution at a given point given its distance vector "d" from the Gaussian mean. More... | |
| template<typename VECTORLIKE1 , typename MATRIXLIKE1 , typename VECTORLIKE2 , typename MATRIXLIKE2 > | |
| double | mrpt::math::KLD_Gaussians (const VECTORLIKE1 &mu0, const MATRIXLIKE1 &cov0, const VECTORLIKE2 &mu1, const MATRIXLIKE2 &cov1) |
| Kullback-Leibler divergence (KLD) between two independent multivariate Gaussians. More... | |
| double | mrpt::math::normalQuantile (double p) |
| Evaluates the Gaussian distribution quantile for the probability value p=[0,1]. More... | |
| double | mrpt::math::normalCDF (double p) |
| Evaluates the Gaussian cumulative density function. More... | |
| double | mrpt::math::chi2inv (double P, unsigned int dim=1) |
| The "quantile" of the Chi-Square distribution, for dimension "dim" and probability 0<P<1 (the inverse of chi2CDF) An aproximation from the Wilson-Hilferty transformation is used. More... | |
| double | mrpt::math::noncentralChi2CDF (unsigned int degreesOfFreedom, double noncentrality, double arg) |
| double | mrpt::math::chi2CDF (unsigned int degreesOfFreedom, double arg) |
| double | mrpt::math::chi2PDF (unsigned int degreesOfFreedom, double arg, double accuracy=1e-7) |
| std::pair< double, double > | mrpt::math::noncentralChi2PDF_CDF (unsigned int degreesOfFreedom, double noncentrality, double arg, double eps=1e-7) |
| Returns the 'exact' PDF (first) and CDF (second) of a Non-central chi-squared probability distribution, using an iterative method. More... | |
| template<typename CONTAINER , typename T > | |
| void | mrpt::math::confidenceIntervals (const CONTAINER &data, T &out_mean, T &out_lower_conf_interval, T &out_upper_conf_interval, const double confidenceInterval=0.1, const size_t histogramNumBins=1000) |
| Return the mean and the 10%-90% confidence points (or with any other confidence value) of a set of samples by building the cummulative CDF of all the elements of the container. More... | |
| std::string | mrpt::math::MATLAB_plotCovariance2D (const CMatrixFloat &cov22, const CVectorFloat &mean, float stdCount, const std::string &style=std::string("b"), size_t nEllipsePoints=30) |
| Generates a string with the MATLAB commands required to plot an confidence interval (ellipse) for a 2D Gaussian ('float' version). More... | |
| std::string | mrpt::math::MATLAB_plotCovariance2D (const CMatrixDouble &cov22, const CVectorDouble &mean, float stdCount, const std::string &style=std::string("b"), size_t nEllipsePoints=30) |
| Generates a string with the MATLAB commands required to plot an confidence interval (ellipse) for a 2D Gaussian ('double' version). More... | |
Probability density distributions (pdf) distance metrics | |
| template<class VECTORLIKE1 , class VECTORLIKE2 , class MAT > | |
| MAT::Scalar | mrpt::math::mahalanobisDistance2 (const VECTORLIKE1 &X, const VECTORLIKE2 &MU, const MAT &COV) |
| Computes the squared mahalanobis distance of a vector X given the mean MU and the covariance inverse COV_inv
. More... | |
| template<class VECTORLIKE1 , class VECTORLIKE2 , class MAT > | |
| VECTORLIKE1::Scalar | mrpt::math::mahalanobisDistance (const VECTORLIKE1 &X, const VECTORLIKE2 &MU, const MAT &COV) |
| Computes the mahalanobis distance of a vector X given the mean MU and the covariance inverse COV_inv
. More... | |
| template<class VECTORLIKE , class MAT1 , class MAT2 , class MAT3 > | |
| MAT1::Scalar | mrpt::math::mahalanobisDistance2 (const VECTORLIKE &mean_diffs, const MAT1 &COV1, const MAT2 &COV2, const MAT3 &CROSS_COV12) |
| Computes the squared mahalanobis distance between two non-independent Gaussians, given the two covariance matrices and the vector with the difference of their means. More... | |
| template<class VECTORLIKE , class MAT1 , class MAT2 , class MAT3 > | |
| VECTORLIKE::Scalar | mrpt::math::mahalanobisDistance (const VECTORLIKE &mean_diffs, const MAT1 &COV1, const MAT2 &COV2, const MAT3 &CROSS_COV12) |
| Computes the mahalanobis distance between two non-independent Gaussians (or independent if CROSS_COV12=nullptr), given the two covariance matrices and the vector with the difference of their means. More... | |
| template<class VECTORLIKE , class MATRIXLIKE > | |
| MATRIXLIKE::Scalar | mrpt::math::mahalanobisDistance2 (const VECTORLIKE &delta_mu, const MATRIXLIKE &cov) |
| Computes the squared mahalanobis distance between a point and a Gaussian, given the covariance matrix and the vector with the difference between the mean and the point. More... | |
| template<class VECTORLIKE , class MATRIXLIKE > | |
| MATRIXLIKE::Scalar | mrpt::math::mahalanobisDistance (const VECTORLIKE &delta_mu, const MATRIXLIKE &cov) |
| Computes the mahalanobis distance between a point and a Gaussian, given the covariance matrix and the vector with the difference between the mean and the point. More... | |
| template<typename T > | |
| T | mrpt::math::productIntegralTwoGaussians (const std::vector< T > &mean_diffs, const CMatrixDynamic< T > &COV1, const CMatrixDynamic< T > &COV2) |
| Computes the integral of the product of two Gaussians, with means separated by "mean_diffs" and covariances "COV1" and "COV2". More... | |
| template<typename T , size_t DIM> | |
| T | mrpt::math::productIntegralTwoGaussians (const std::vector< T > &mean_diffs, const CMatrixFixed< T, DIM, DIM > &COV1, const CMatrixFixed< T, DIM, DIM > &COV2) |
| Computes the integral of the product of two Gaussians, with means separated by "mean_diffs" and covariances "COV1" and "COV2". More... | |
| template<typename T , class VECLIKE , class MATLIKE1 , class MATLIKE2 > | |
| void | mrpt::math::productIntegralAndMahalanobisTwoGaussians (const VECLIKE &mean_diffs, const MATLIKE1 &COV1, const MATLIKE2 &COV2, T &maha2_out, T &intprod_out, const MATLIKE1 *CROSS_COV12=nullptr) |
| Computes both, the integral of the product of two Gaussians and their square Mahalanobis distance. More... | |
| template<typename T , class VECLIKE , class MATRIXLIKE > | |
| void | mrpt::math::mahalanobisDistance2AndLogPDF (const VECLIKE &diff_mean, const MATRIXLIKE &cov, T &maha2_out, T &log_pdf_out) |
| Computes both, the logarithm of the PDF and the square Mahalanobis distance between a point (given by its difference wrt the mean) and a Gaussian. More... | |
| template<typename T , class VECLIKE , class MATRIXLIKE > | |
| void | mrpt::math::mahalanobisDistance2AndPDF (const VECLIKE &diff_mean, const MATRIXLIKE &cov, T &maha2_out, T &pdf_out) |
| Computes both, the PDF and the square Mahalanobis distance between a point (given by its difference wrt the mean) and a Gaussian. More... | |
| template<class VECTOR_OF_VECTORS , class MATRIXLIKE , class VECTORLIKE , class VECTORLIKE2 , class VECTORLIKE3 > | |
| void | mrpt::math::covariancesAndMeanWeighted (const VECTOR_OF_VECTORS &elements, MATRIXLIKE &covariances, VECTORLIKE &means, const VECTORLIKE2 *weights_mean, const VECTORLIKE3 *weights_cov, const bool *elem_do_wrap2pi=nullptr) |
| Computes covariances and mean of any vector of containers, given optional weights for the different samples. More... | |
| template<class VECTOR_OF_VECTORS , class MATRIXLIKE , class VECTORLIKE > | |
| void | mrpt::math::covariancesAndMean (const VECTOR_OF_VECTORS &elements, MATRIXLIKE &covariances, VECTORLIKE &means, const bool *elem_do_wrap2pi=nullptr) |
| Computes covariances and mean of any vector of containers. More... | |
| template<class VECTORLIKE1 , class VECTORLIKE2 > | |
| void | mrpt::math::weightedHistogram (const VECTORLIKE1 &values, const VECTORLIKE1 &weights, float binWidth, VECTORLIKE2 &out_binCenters, VECTORLIKE2 &out_binValues) |
| Computes the weighted histogram for a vector of values and their corresponding weights. More... | |
| template<class VECTORLIKE1 , class VECTORLIKE2 > | |
| void | mrpt::math::weightedHistogramLog (const VECTORLIKE1 &values, const VECTORLIKE1 &log_weights, float binWidth, VECTORLIKE2 &out_binCenters, VECTORLIKE2 &out_binValues) |
| Computes the weighted histogram for a vector of values and their corresponding log-weights. More... | |
| double | mrpt::math::averageLogLikelihood (const CVectorDouble &logLikelihoods) |
| A numerically-stable method to compute average likelihood values with strongly different ranges (unweighted likelihoods: compute the arithmetic mean). More... | |
| double | mrpt::math::averageWrap2Pi (const CVectorDouble &angles) |
Computes the average of a sequence of angles in radians taking into account the correct wrapping in the range ![$ ]-\pi,\pi [ $](form_51.png) , for example, the mean of (2,-2) is , for example, the mean of (2,-2) is 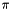 , not 0. More... , not 0. More... | |
| double | mrpt::math::averageLogLikelihood (const CVectorDouble &logWeights, const CVectorDouble &logLikelihoods) |
| A numerically-stable method to average likelihood values with strongly different ranges (weighted likelihoods). More... | |
Function Documentation
◆ averageLogLikelihood() [1/2]
| double mrpt::math::averageLogLikelihood | ( | const CVectorDouble & | logLikelihoods | ) |
A numerically-stable method to compute average likelihood values with strongly different ranges (unweighted likelihoods: compute the arithmetic mean).
This method implements this equation:
![\[ return = - \log N + \log \sum_{i=1}^N e^{ll_i-ll_{max}} + ll_{max} \]](form_50.png)
See also the tutorial page.
Definition at line 293 of file math.cpp.
References mrpt::math::maximum(), MRPT_CHECK_NORMAL_NUMBER, MRPT_END, MRPT_START, mrpt::math::CVectorDynamic< T >::size(), and THROW_EXCEPTION.
Referenced by mrpt::maps::CBeaconMap::internal_computeObservationLikelihood(), mrpt::slam::PF_implementation< mrpt::math::TPose3D, CMonteCarloLocalization3D, mrpt::bayes::particle_storage_mode::VALUE >::PF_SLAM_particlesEvaluator_AuxPFOptimal(), and mrpt::slam::PF_implementation< mrpt::math::TPose3D, CMonteCarloLocalization3D, mrpt::bayes::particle_storage_mode::VALUE >::PF_SLAM_particlesEvaluator_AuxPFStandard().
◆ averageLogLikelihood() [2/2]
| double mrpt::math::averageLogLikelihood | ( | const CVectorDouble & | logWeights, |
| const CVectorDouble & | logLikelihoods | ||
| ) |
A numerically-stable method to average likelihood values with strongly different ranges (weighted likelihoods).
This method implements this equation:
![\[ return = \log \left( \frac{1}{\sum_i e^{lw_i}} \sum_i e^{lw_i} e^{ll_i} \right) \]](form_53.png)
See also the tutorial page.
Definition at line 261 of file math.cpp.
References ASSERT_, mrpt::math::CVectorDynamic< T >::begin(), mrpt::math::CVectorDynamic< T >::end(), mrpt::math::maximum(), MRPT_CHECK_NORMAL_NUMBER, MRPT_END, MRPT_START, mrpt::math::CVectorDynamic< T >::size(), and THROW_EXCEPTION.
◆ averageWrap2Pi()
| double mrpt::math::averageWrap2Pi | ( | const CVectorDouble & | angles | ) |
Computes the average of a sequence of angles in radians taking into account the correct wrapping in the range ![$ ]-\pi,\pi [ $](form_51.png) , for example, the mean of (2,-2) is
, for example, the mean of (2,-2) is 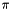 , not 0.
, not 0.
Definition at line 315 of file math.cpp.
References mrpt::math::CVectorDynamic< T >::empty(), M_2PI, M_PI, and mrpt::math::CVectorDynamic< T >::size().
Referenced by mrpt::topography::path_from_rtk_gps().
◆ chi2CDF()
| double mrpt::math::chi2CDF | ( | unsigned int | degreesOfFreedom, |
| double | arg | ||
| ) |
Cumulative chi square distribution.
Computes the cumulative density of a chi square distribution with degreesOfFreedom and tolerance accuracy at the given argument arg, i.e. the probability that a random number drawn from the distribution is below arg by calling noncentralChi2CDF(degreesOfFreedom, 0.0, arg, accuracy).
- Note
- Function code from the Vigra project (http://hci.iwr.uni-heidelberg.de/vigra/); code under "MIT X11 License", GNU GPL-compatible.
Definition at line 505 of file math.cpp.
References mrpt::math::noncentralChi2CDF().
◆ chi2inv()
| double mrpt::math::chi2inv | ( | double | P, |
| unsigned int | dim = 1 |
||
| ) |
The "quantile" of the Chi-Square distribution, for dimension "dim" and probability 0<P<1 (the inverse of chi2CDF) An aproximation from the Wilson-Hilferty transformation is used.
- Note
- Equivalent to MATLAB chi2inv(), but note that this is just an approximation, which becomes very poor for small values of "P".
Definition at line 42 of file math.cpp.
References ASSERT_, and mrpt::math::normalQuantile().
Referenced by mrpt::slam::CGridMapAligner::AlignPDF_robustMatch(), mrpt::slam::data_association_full_covariance(), mrpt::hmtslam::CHMTSLAM::LSLAM_process_message_from_TBI(), mrpt::slam::PF_implementation< mrpt::math::TPose3D, CMonteCarloLocalization3D, mrpt::bayes::particle_storage_mode::VALUE >::PF_SLAM_implementation_pfAuxiliaryPFStandardAndOptimal(), mrpt::slam::PF_implementation< mrpt::math::TPose3D, CMonteCarloLocalization3D, mrpt::bayes::particle_storage_mode::VALUE >::PF_SLAM_implementation_pfStandardProposal(), mrpt::tfest::se2_l2_robust(), and TEST().
◆ chi2PDF()
| double mrpt::math::chi2PDF | ( | unsigned int | degreesOfFreedom, |
| double | arg, | ||
| double | accuracy = 1e-7 |
||
| ) |
Chi square distribution PDF. Computes the density of a chi square distribution with degreesOfFreedom and tolerance accuracy at the given argument arg by calling noncentralChi2(degreesOfFreedom, 0.0, arg, accuracy).
- Note
- Function code from the Vigra project (http://hci.iwr.uni-heidelberg.de/vigra/); code under "MIT X11 License", GNU GPL-compatible.
- Equivalent to MATLAB's chi2pdf(arg,degreesOfFreedom)
Definition at line 595 of file math.cpp.
References mrpt::math::noncentralChi2PDF_CDF().
Referenced by TEST().
◆ confidenceIntervals()
| void mrpt::math::confidenceIntervals | ( | const CONTAINER & | data, |
| T & | out_mean, | ||
| T & | out_lower_conf_interval, | ||
| T & | out_upper_conf_interval, | ||
| const double | confidenceInterval = 0.1, |
||
| const size_t | histogramNumBins = 1000 |
||
| ) |
Return the mean and the 10%-90% confidence points (or with any other confidence value) of a set of samples by building the cummulative CDF of all the elements of the container.
The container can be any MRPT container (CArray, matrices, vectors).
- Parameters
-
confidenceInterval A number in the range (0,1) such as the confidence interval will be [100*confidenceInterval, 100*(1-confidenceInterval)].
Definition at line 216 of file distributions.h.
References ASSERT_, mrpt::math::cumsum(), mrpt::opengl::internal::data, mrpt::math::distance(), mrpt::math::histogram(), mrpt::math::maximum(), mrpt::math::mean(), MRPT_END, and MRPT_START.
Referenced by mrpt::apps::MonteCarloLocalization_Base::do_pf_localization().
◆ covariancesAndMean()
| void mrpt::math::covariancesAndMean | ( | const VECTOR_OF_VECTORS & | elements, |
| MATRIXLIKE & | covariances, | ||
| VECTORLIKE & | means, | ||
| const bool * | elem_do_wrap2pi = nullptr |
||
| ) |
Computes covariances and mean of any vector of containers.
- Parameters
-
elements Any kind of vector of vectors/arrays, eg. std::vector<mrpt::math::CVectorDouble>, with all the input samples, each sample in a "row". covariances Output estimated covariance; it can be a fixed/dynamic matrix or a matrixview. means Output estimated mean; it can be CVectorDouble/CVectorFixedDouble, etc... elem_do_wrap2pi If !=nullptr; it must point to an array of "bool" of size()==dimension of each element, stating if it's needed to do a wrap to [-pi,pi] to each dimension.
Definition at line 381 of file data_utils.h.
References mrpt::math::covariancesAndMeanWeighted().
Referenced by mrpt::math::getRegressionLine(), mrpt::math::getRegressionPlane(), and mrpt::math::transform_gaussian_montecarlo().
◆ covariancesAndMeanWeighted()
|
inline |
Computes covariances and mean of any vector of containers, given optional weights for the different samples.
- Parameters
-
elements Any kind of vector of vectors/arrays, eg. std::vector<mrpt::math::CVectorDouble>, with all the input samples, each sample in a "row". covariances Output estimated covariance; it can be a fixed/dynamic matrix or a matrixview. means Output estimated mean; it can be CVectorDouble/CVectorFixedDouble, etc... weights_mean If !=nullptr, it must point to a vector of size()==number of elements, with normalized weights to take into account for the mean. weights_cov If !=nullptr, it must point to a vector of size()==number of elements, with normalized weights to take into account for the covariance. elem_do_wrap2pi If !=nullptr; it must point to an array of "bool" of size()==dimension of each element, stating if it's needed to do a wrap to [-pi,pi] to each dimension.
- See also
- This method is used in mrpt::math::unscented_transform_gaussian
Definition at line 261 of file data_utils.h.
References ASSERTDEB_, ASSERTMSG_, M_2PI, M_PI, and mrpt::math::wrapToPi().
Referenced by mrpt::math::covariancesAndMean(), and mrpt::math::transform_gaussian_unscented().
◆ KLD_Gaussians()
| double mrpt::math::KLD_Gaussians | ( | const VECTORLIKE1 & | mu0, |
| const MATRIXLIKE1 & | cov0, | ||
| const VECTORLIKE2 & | mu1, | ||
| const MATRIXLIKE2 & | cov1 | ||
| ) |
Kullback-Leibler divergence (KLD) between two independent multivariate Gaussians.
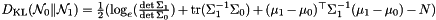
Definition at line 107 of file distributions.h.
References ASSERT_, MRPT_END, MRPT_START, and mrpt::math::multiply_HCHt_scalar().
◆ mahalanobisDistance() [1/3]
|
inline |
Computes the mahalanobis distance of a vector X given the mean MU and the covariance inverse COV_inv
![\[ d = \sqrt{ (X-MU)^\top \Sigma^{-1} (X-MU) } \]](form_43.png)
.
Definition at line 57 of file data_utils.h.
References mrpt::math::mahalanobisDistance2().
Referenced by mrpt::math::mahalanobisDistance(), mrpt::poses::CPose3DQuatPDFGaussian::mahalanobisDistanceTo(), and mrpt::topography::path_from_rtk_gps().
◆ mahalanobisDistance() [2/3]
|
inline |
Computes the mahalanobis distance between two non-independent Gaussians (or independent if CROSS_COV12=nullptr), given the two covariance matrices and the vector with the difference of their means.
![\[ d = \sqrt{ \Delta_\mu^\top (\Sigma_1 + \Sigma_2 - 2 \Sigma_12 )^{-1} \Delta_\mu } \]](form_45.png)
Definition at line 97 of file data_utils.h.
References mrpt::math::mahalanobisDistance().
◆ mahalanobisDistance() [3/3]
|
inline |
Computes the mahalanobis distance between a point and a Gaussian, given the covariance matrix and the vector with the difference between the mean and the point.
![\[ d^2 = \sqrt( \Delta_\mu^\top \Sigma^{-1} \Delta_\mu ) \]](form_47.png)
Definition at line 124 of file data_utils.h.
References mrpt::math::cov(), and mrpt::math::mahalanobisDistance2().
◆ mahalanobisDistance2() [1/3]
| MAT::Scalar mrpt::math::mahalanobisDistance2 | ( | const VECTORLIKE1 & | X, |
| const VECTORLIKE2 & | MU, | ||
| const MAT & | COV | ||
| ) |
Computes the squared mahalanobis distance of a vector X given the mean MU and the covariance inverse COV_inv
![\[ d^2 = (X-MU)^\top \Sigma^{-1} (X-MU) \]](form_42.png)
.
Definition at line 35 of file data_utils.h.
References ASSERT_, mrpt::math::MatrixVectorBase< T, CVectorDynamic< T > >::dot(), MRPT_END, and MRPT_START.
Referenced by mrpt::math::mahalanobisDistance(), and mrpt::graphslam::deciders::CLoopCloserERD< GRAPH_T >::mahalanobisDistanceOdometryToICPEdge().
◆ mahalanobisDistance2() [2/3]
| MAT1::Scalar mrpt::math::mahalanobisDistance2 | ( | const VECTORLIKE & | mean_diffs, |
| const MAT1 & | COV1, | ||
| const MAT2 & | COV2, | ||
| const MAT3 & | CROSS_COV12 | ||
| ) |
Computes the squared mahalanobis distance between two non-independent Gaussians, given the two covariance matrices and the vector with the difference of their means.
![\[ d^2 = \Delta_\mu^\top (\Sigma_1 + \Sigma_2 - 2 \Sigma_12 )^{-1} \Delta_\mu \]](form_44.png)
Definition at line 70 of file data_utils.h.
References ASSERT_, MRPT_END, MRPT_START, and mrpt::math::multiply_HCHt_scalar().
◆ mahalanobisDistance2() [3/3]
|
inline |
Computes the squared mahalanobis distance between a point and a Gaussian, given the covariance matrix and the vector with the difference between the mean and the point.
![\[ d^2 = \Delta_\mu^\top \Sigma^{-1} \Delta_\mu \]](form_46.png)
Definition at line 110 of file data_utils.h.
References ASSERTDEB_, mrpt::math::CMatrixDynamic< T >::cols(), mrpt::math::cov(), mrpt::math::MatrixBase< Scalar, Derived >::inverse(), mrpt::math::MatrixVectorBase< Scalar, Derived >::isSquare(), and mrpt::math::multiply_HtCH_scalar().
◆ mahalanobisDistance2AndLogPDF()
| void mrpt::math::mahalanobisDistance2AndLogPDF | ( | const VECLIKE & | diff_mean, |
| const MATRIXLIKE & | cov, | ||
| T & | maha2_out, | ||
| T & | log_pdf_out | ||
| ) |
Computes both, the logarithm of the PDF and the square Mahalanobis distance between a point (given by its difference wrt the mean) and a Gaussian.
Definition at line 209 of file data_utils.h.
References ASSERTDEB_, mrpt::math::CMatrixDynamic< T >::cols(), mrpt::math::cov(), mrpt::math::MatrixBase< Scalar, Derived >::det(), mrpt::math::MatrixBase< Scalar, Derived >::inverse_LLt(), mrpt::math::MatrixVectorBase< Scalar, Derived >::isSquare(), M_2PI, MRPT_END, MRPT_START, and mrpt::math::multiply_HtCH_scalar().
Referenced by mrpt::slam::data_association_full_covariance(), mrpt::math::mahalanobisDistance2AndPDF(), mrpt::slam::CRangeBearingKFSLAM2D::OnGetObservationsAndDataAssociation(), and TEST().
◆ mahalanobisDistance2AndPDF()
|
inline |
Computes both, the PDF and the square Mahalanobis distance between a point (given by its difference wrt the mean) and a Gaussian.
Definition at line 231 of file data_utils.h.
References mrpt::math::cov(), and mrpt::math::mahalanobisDistance2AndLogPDF().
◆ MATLAB_plotCovariance2D() [1/2]
| string mrpt::math::MATLAB_plotCovariance2D | ( | const CMatrixFloat & | cov22, |
| const CVectorFloat & | mean, | ||
| float | stdCount, | ||
| const std::string & | style = std::string("b"), |
||
| size_t | nEllipsePoints = 30 |
||
| ) |
Generates a string with the MATLAB commands required to plot an confidence interval (ellipse) for a 2D Gaussian ('float' version).
- Parameters
-
cov22 The 2x2 covariance matrix mean The 2-length vector with the mean stdCount How many "quantiles" to get into the area of the ellipse: 2: 95%, 3:99.97%,... style A matlab style string, for colors, line styles,... nEllipsePoints The number of points in the ellipse to generate
Definition at line 356 of file math.cpp.
References mrpt::math::cov(), mrpt::math::mean(), MRPT_END, and MRPT_START.
Referenced by mrpt::slam::CRangeBearingKFSLAM2D::saveMapAndPath2DRepresentationAsMATLABFile(), and mrpt::slam::CRangeBearingKFSLAM::saveMapAndPath2DRepresentationAsMATLABFile().
◆ MATLAB_plotCovariance2D() [2/2]
| string mrpt::math::MATLAB_plotCovariance2D | ( | const CMatrixDouble & | cov22, |
| const CVectorDouble & | mean, | ||
| float | stdCount, | ||
| const std::string & | style = std::string("b"), |
||
| size_t | nEllipsePoints = 30 |
||
| ) |
Generates a string with the MATLAB commands required to plot an confidence interval (ellipse) for a 2D Gaussian ('double' version).
- Parameters
-
cov22 The 2x2 covariance matrix mean The 2-length vector with the mean stdCount How many "quantiles" to get into the area of the ellipse: 2: 95%, 3:99.97%,... style A matlab style string, for colors, line styles,... nEllipsePoints The number of points in the ellipse to generate
Definition at line 372 of file math.cpp.
References mrpt::math::MatrixVectorBase< T, CMatrixFixed< T, ROWS, COLS > >::array(), mrpt::math::CMatrixFixed< T, ROWS, COLS >::asEigen(), mrpt::math::CMatrixDynamic< T >::asEigen(), ASSERT_, mrpt::math::CMatrixDynamic< T >::cols(), mrpt::math::cov(), mrpt::math::MatrixBase< Scalar, Derived >::eig(), mrpt::format(), M_2PI, mrpt::math::mean(), MRPT_END_WITH_CLEAN_UP, MRPT_START, mrpt::math::CMatrixDynamic< T >::rows(), mrpt::math::MatrixBase< T, CMatrixFixed< T, ROWS, COLS > >::setDiagonal(), and mrpt::math::MatrixVectorBase< T, CMatrixFixed< T, ROWS, COLS > >::transpose().
◆ noncentralChi2CDF()
| double mrpt::math::noncentralChi2CDF | ( | unsigned int | degreesOfFreedom, |
| double | noncentrality, | ||
| double | arg | ||
| ) |
Cumulative non-central chi square distribution (approximate).
Computes approximate values of the cumulative density of a chi square distribution with degreesOfFreedom, and noncentrality parameter noncentrality at the given argument arg, i.e. the probability that a random number drawn from the distribution is below arg It uses the approximate transform into a normal distribution due to Wilson and Hilferty (see Abramovitz, Stegun: "Handbook of Mathematical Functions", formula 26.3.32). The algorithm's running time is independent of the inputs. The accuracy is only about 0.1 for few degrees of freedom, but reaches about 0.001 above dof = 5.
- Note
- Function code from the Vigra project (http://hci.iwr.uni-heidelberg.de/vigra/); code under "MIT X11 License", GNU GPL-compatible.
- See also
- noncentralChi2PDF_CDF
Definition at line 603 of file math.cpp.
References mrpt::square().
Referenced by mrpt::math::chi2CDF().
◆ noncentralChi2PDF_CDF()
| std::pair< double, double > mrpt::math::noncentralChi2PDF_CDF | ( | unsigned int | degreesOfFreedom, |
| double | noncentrality, | ||
| double | arg, | ||
| double | eps = 1e-7 |
||
| ) |
Returns the 'exact' PDF (first) and CDF (second) of a Non-central chi-squared probability distribution, using an iterative method.
- Note
- Equivalent to MATLAB's ncx2cdf(arg,degreesOfFreedom,noncentrality)
Definition at line 528 of file math.cpp.
References ASSERTMSG_, mrpt::obs::gnss::b1, eps, noncentralChi2OneIteration(), mrpt::math::sum(), and THROW_EXCEPTION.
Referenced by mrpt::math::chi2PDF(), and TEST().
◆ normalCDF()
| double mrpt::math::normalCDF | ( | double | p | ) |
Evaluates the Gaussian cumulative density function.
The employed approximation is that from W. J. Cody freely available in http://www.netlib.org/specfun/erf
- Note
- Equivalent to MATLAB normcdf(x,mu,s) with p=(x-mu)/s
Definition at line 134 of file math.cpp.
References ASSERT_.
Referenced by mrpt::math::normalQuantile(), and TEST().
◆ normalPDF() [1/3]
| double mrpt::math::normalPDF | ( | double | x, |
| double | mu, | ||
| double | std | ||
| ) |
Evaluates the univariate normal (Gaussian) distribution at a given point "x".
Definition at line 33 of file math.cpp.
References mrpt::square().
Referenced by mrpt::poses::CPose3DQuatPDFGaussian::evaluateNormalizedPDF(), mrpt::poses::CPosePDFGaussian::evaluateNormalizedPDF(), mrpt::poses::CPosePDFGaussianInf::evaluateNormalizedPDF(), mrpt::poses::CPosePDFSOG::evaluateNormalizedPDF(), mrpt::poses::CPose3DQuatPDFGaussian::evaluatePDF(), mrpt::poses::CPosePDFGaussian::evaluatePDF(), mrpt::poses::CPosePDFGaussianInf::evaluatePDF(), mrpt::poses::CPointPDFSOG::evaluatePDF(), mrpt::poses::CPosePDFSOG::evaluatePDF(), mrpt::poses::CPosePDFParticles::evaluatePDF_parzen(), and TEST().
◆ normalPDF() [2/3]
|
inline |
Evaluates the multivariate normal (Gaussian) distribution at a given point "x".
- Parameters
-
x A vector or column or row matrix with the point at which to evaluate the pdf. mu A vector or column or row matrix with the Gaussian mean. cov The covariance matrix of the Gaussian. scaled_pdf If set to true, the PDF will be scaled to be in the range [0,1], in contrast to its integral from [-inf,+inf] being 1.
Definition at line 70 of file distributions.h.
References mrpt::math::cov(), mrpt::math::MatrixBase< Scalar, Derived >::inverse(), and mrpt::math::normalPDFInf().
◆ normalPDF() [3/3]
| MATRIXLIKE::Scalar mrpt::math::normalPDF | ( | const VECTORLIKE & | d, |
| const MATRIXLIKE & | cov | ||
| ) |
Evaluates the multivariate normal (Gaussian) distribution at a given point given its distance vector "d" from the Gaussian mean.
Definition at line 81 of file distributions.h.
References ASSERTDEB_, mrpt::math::CMatrixDynamic< T >::cols(), mrpt::math::cov(), mrpt::math::MatrixBase< Scalar, Derived >::det(), mrpt::math::MatrixBase< Scalar, Derived >::inverse_LLt(), mrpt::math::MatrixVectorBase< Scalar, Derived >::isSquare(), M_2PI, MRPT_END, MRPT_START, and mrpt::math::multiply_HtCH_scalar().
◆ normalPDFInf()
|
inline |
Evaluates the multivariate normal (Gaussian) distribution at a given point "x".
- Parameters
-
x A vector or column or row matrix with the point at which to evaluate the pdf. mu A vector or column or row matrix with the Gaussian mean. cov_inv The inverse covariance (information) matrix of the Gaussian. scaled_pdf If set to true, the PDF will be scaled to be in the range [0,1], in contrast to its integral from [-inf,+inf] being 1.
Definition at line 38 of file distributions.h.
References ASSERTDEB_, M_2PI, MRPT_END, MRPT_START, and mrpt::math::multiply_HtCH_scalar().
Referenced by mrpt::poses::CPose3DQuatPDFGaussianInf::evaluateNormalizedPDF(), mrpt::poses::CPose3DQuatPDFGaussianInf::evaluatePDF(), and mrpt::math::normalPDF().
◆ normalQuantile()
| double mrpt::math::normalQuantile | ( | double | p | ) |
Evaluates the Gaussian distribution quantile for the probability value p=[0,1].
The employed approximation is that from Peter J. Acklam (pjacklam@online.no), freely available in http://home.online.no/~pjacklam.
Definition at line 80 of file math.cpp.
References ASSERT_, and mrpt::math::normalCDF().
Referenced by mrpt::math::chi2inv().
◆ productIntegralAndMahalanobisTwoGaussians()
| void mrpt::math::productIntegralAndMahalanobisTwoGaussians | ( | const VECLIKE & | mean_diffs, |
| const MATLIKE1 & | COV1, | ||
| const MATLIKE2 & | COV2, | ||
| T & | maha2_out, | ||
| T & | intprod_out, | ||
| const MATLIKE1 * | CROSS_COV12 = nullptr |
||
| ) |
Computes both, the integral of the product of two Gaussians and their square Mahalanobis distance.
Definition at line 180 of file data_utils.h.
References ASSERT_, M_2PI, and mrpt::math::multiply_HCHt_scalar().
◆ productIntegralTwoGaussians() [1/2]
| T mrpt::math::productIntegralTwoGaussians | ( | const std::vector< T > & | mean_diffs, |
| const CMatrixDynamic< T > & | COV1, | ||
| const CMatrixDynamic< T > & | COV2 | ||
| ) |
Computes the integral of the product of two Gaussians, with means separated by "mean_diffs" and covariances "COV1" and "COV2".
![\[ D = \frac{1}{(2 \pi)^{0.5 N} \sqrt{} } \exp( \Delta_\mu^\top (\Sigma_1 + \Sigma_2 - 2 \Sigma_12)^{-1} \Delta_\mu) \]](form_48.png)
Definition at line 136 of file data_utils.h.
References ASSERT_, mrpt::math::MatrixBase< T, CMatrixDynamic< T > >::det(), mrpt::math::MatrixBase< T, CMatrixDynamic< T > >::inverse_LLt(), and M_2PI.
◆ productIntegralTwoGaussians() [2/2]
| T mrpt::math::productIntegralTwoGaussians | ( | const std::vector< T > & | mean_diffs, |
| const CMatrixFixed< T, DIM, DIM > & | COV1, | ||
| const CMatrixFixed< T, DIM, DIM > & | COV2 | ||
| ) |
Computes the integral of the product of two Gaussians, with means separated by "mean_diffs" and covariances "COV1" and "COV2".
![\[ D = \frac{1}{(2 \pi)^{0.5 N} \sqrt{} } \exp( \Delta_\mu^\top (\Sigma_1 + \Sigma_2)^{-1} \Delta_\mu) \]](form_49.png)
Definition at line 159 of file data_utils.h.
References ASSERT_, mrpt::math::MatrixBase< T, CMatrixFixed< T, ROWS, COLS > >::det(), M_2PI, and mrpt::math::UNINITIALIZED_MATRIX.
◆ weightedHistogram()
| void mrpt::math::weightedHistogram | ( | const VECTORLIKE1 & | values, |
| const VECTORLIKE1 & | weights, | ||
| float | binWidth, | ||
| VECTORLIKE2 & | out_binCenters, | ||
| VECTORLIKE2 & | out_binValues | ||
| ) |
Computes the weighted histogram for a vector of values and their corresponding weights.
- Parameters
-
values [IN] The N values weights [IN] The weights for the corresponding N values (don't need to be normalized) binWidth [IN] The desired width of the bins out_binCenters [OUT] The centers of the M bins generated to cover from the minimum to the maximum value of "values" with the given "binWidth" out_binValues [OUT] The ratio of values at each given bin, such as the whole vector sums up the unity.
- See also
- weightedHistogramLog
Definition at line 404 of file data_utils.h.
References ASSERT_, ASSERTDEB_, mrpt::math::maximum(), mrpt::math::minimum(), MRPT_END, MRPT_START, and mrpt::round().
◆ weightedHistogramLog()
| void mrpt::math::weightedHistogramLog | ( | const VECTORLIKE1 & | values, |
| const VECTORLIKE1 & | log_weights, | ||
| float | binWidth, | ||
| VECTORLIKE2 & | out_binCenters, | ||
| VECTORLIKE2 & | out_binValues | ||
| ) |
Computes the weighted histogram for a vector of values and their corresponding log-weights.
- Parameters
-
values [IN] The N values weights [IN] The log-weights for the corresponding N values (don't need to be normalized) binWidth [IN] The desired width of the bins out_binCenters [OUT] The centers of the M bins generated to cover from the minimum to the maximum value of "values" with the given "binWidth" out_binValues [OUT] The ratio of values at each given bin, such as the whole vector sums up the unity.
- See also
- weightedHistogram
Definition at line 461 of file data_utils.h.
References ASSERT_, ASSERTDEB_, mrpt::math::maximum(), mrpt::math::minimum(), MRPT_END, MRPT_START, and mrpt::round().